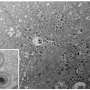
The newly discovered receptor for enterovirus D68, MFSD6, is partially localized to the cell surface where it can facilitate viral entry. Cells are shown that have the cell nucleus in the middle, colored in blue, and the cell surface, colored in magenta. MFSD6 (in green) is present in dot-like structures, some of which are present at the cell surface (white dots indicated with arrows). Credit: Lily Xu and Christine Peters/Stanford University
Researchers found a protein that's essential for an enterovirus to enter human cells. Although not the infamous exampleÔÇöthat title goes to poliovirusÔÇöother enteroviruses such as enterovirus D68 can cause similar paralytic symptoms in young children.
Indeed, small enterovirus D68 outbreaks have occurred in the U.S., but there's no vaccine or specific treatment.
Now, writing in the journal , researchers at the Stanford University School of Medicine, the Department of Energy's SLAC National Accelerator Laboratory, and three other institutions have identified a key to enterovirus D68 infection, which could help shape future vaccinations and treatments.
That key sits on the surface of human cells. Viruses must bind to cells' receptor proteins to gain entry and start infection. Researchers in Stanford professor Jan Carette's lab uncovered which receptor protein unlocks the door to the cell for enterovirus D68 by turning off the expression of each gene in the entire human genome one by one.
"It's an unbiased way of understanding which receptor is the most important for the virus out of the 20,000 proteins," said Carette.
The result: Enterovirus D68 could not enter cells when a protein called MFSD6 was absent. What's more, the team found that mice injected with a soluble form of the MFSD6 protein were almost completely protected from infection. The soluble form acted as a decoy receptor, coating the virus to block it from binding with actual cell receptors.
"Once you put the key in, no one else can put their key in the same spot," said Lily Xu, Ph.D. candidate at Stanford and co-first author of the results.
To design vaccines capable of tightly binding to the virus and blocking infection, researchers need to better understand how the virus and MFSD6 receptor protein fit together.
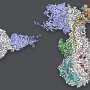
In SLAC and Stanford professor Wah Chiu's lab, a team used SLAC's state-of-the-art cryogenic electron microscopy (cryo-EM) facilities to magnify the virus and protein by a factor of over 100,000. Combining this high-resolution imaging technique with the modeling skills of Stanford research scientist Grigore Pintilie, the team created a 3D model of the interaction between the virus and protein at the molecular level.
"These 3D structures are useful because they can help design other proteins or molecules that get in there to disturb the whole infection cycle," Pintilie said.
Unexpectedly, cryo-EM revealed multiple sugars attached to the receptor protein. Stanford chemistry professor Carolyn Bertozzi and her postdoctoral fellow David Roberts used mass spectrometry to identify these sugars, which could further guide the design of vaccines.
"My dream is that our discovery can lead to the production of host-targeted therapeutics or the development of a drug or antibody that can target enterovirus D68's receptor binding site," said Lauren Varanese, Ph.D. candidate at Stanford and co-first author of this work.
While any potential preventatives or therapeutics based on these results wouldn't be available for five to ten years or more, in the meantime, the structural characterization methods in this work could be extended to other viruses.
"At SLAC, we demonstrated that we have imaging capabilities to retrieve structural details that can help with any emergent virus," said Chiu.
More information: Lauren Varanese et al, MFSD6 is an entry receptor for enterovirus D68, Nature (2025).
Journal information: Nature
Provided by SLAC National Accelerator Laboratory