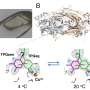
Comparing cryogenic structures with room-temperature samples can help identify errors in computational models

About 95% of all crystal structures obtained for various proteins and deposited in public databases are captured using cryogenic technology. This technology requires frozen conditions. Scientists at St. Jude Children's Research Hospital compared cryogenic structures with those observed at room temperature. The findings, published today in Chemical Science, indicate that freezing can introduce errors, cause certain conformations (shapes) to be missed and lead to inaccuracies in computational models.
Protein structures are essential to the drug development process because they provide a map for how targeted drugs should be designed.
"We need to rethink how we collect, analyze and utilize structural information when we set out to discover bioactive molecules," said corresponding author Marcus Fischer, Ph.D., St. Jude Department of Chemical Biology and Therapeutics. "You can view temperature as an experimental knob we can turn to explore hidden protein conformations."
Temperature makes all the difference
The researchers have shown that freezing distorts the conformations that proteins take, often introducing errors in structures. The team also found that some conformations occurring at room-temperature conditions can be missed if only looking at results from cryogenic techniques.
The researchers conducted a systematic evaluation of cryogenic structures, starting with the T4 lysozyme L99A cavity. This protein is considered a "workhorse" in structural biology for understanding protein stability, rigidity and ligand-binding thermodynamics. Shifting to room-temperature revealed new structural changes that have been missed for decades.
The team tested four additional classes of proteins. The results held true regardless of which type of protein was evaluated.
"When you go out in the winter and are cold, you compress and shrink in on yourself, and in the sun when you're warm you stretch out. Proteins do the same," Fischer said.
Avoiding errors
Computational methods are algorithms that researchers use to make predictions or evaluate data obtained from their experiments. The results indicate that when these methods are built on data from cryogenic structures, errors can be introduced that may taint future results.
Cryogenic techniques have long been favored because they make it easier to obtain the structures. Getting structures at room temperature is more tedious. Although there are ways to mitigate these issues, factors such as data completeness and radiation damage are additional hurdles for many researchers in obtaining room-temperature structures.
While detecting a hidden protein shape is informative, showing the new shape's impact on drug discovery protocols was still missing.
"We saw that the protein adopted a state to interact with ligands, and that missing information may help improve the accuracy of virtual drug screening and protein-ligand interaction simulations," said co-first author Shanshan Bradford, Ph.D., St. Jude Department of Chemical Biology and Therapeutics.
The researchers underscore that when just considering cryogenic structures, there is no way to tell if there are errors, but that comparison with room-temperature structures may help clarify information and potentially reveal additional insights that are otherwise missed.
More information: Shanshan Y. C. Bradford et al, Temperature artifacts in protein structures bias ligand-binding predictions, Chemical Science (2021).
Journal information: Chemical Science
Provided by St. Jude Children's Research Hospital