Study reveals key aspect of the finely tuned regulation of gene expression
Your skin cells are clearly different from your brain cells even though they both develop in the same person and carry the same genes. They are different because each cell type expresses a particular set of genes that is different from the ones expressed by the other. This is possible thanks to cellular mechanisms that tightly regulate gene expression.
In a study published in the Proceedings of the National Academy of Sciences, researchers at Baylor College of Medicine in Dr. Bert O'Malley's group unveil a novel key aspect of the mechanism of gene expression regulation. The findings not only contribute to a better understanding of this essential biological process but also open new possibilities to study alterations in gene expression regulation that lead to disease.
"Gene expression is controlled at different levels," said lead author Dr. Anil K. Panigrahi, assistant professor in the Department of Molecular and Cellular Biology at Baylor. "In this study, we focused on enhancers, one of the critical components that regulate gene expression. Enhancers are segments of DNA that activate gene expression by interacting with the gene's promoter. Enhancers and promoters form physical contact, which imparts the message to the cell of when to express the gene and how much."
Although enhancers and promoters appear to coordinate their actions, it is unclear how this happens. In this study, Panigrahi and his colleagues propose a mechanism that explains the tight connection between enhancers and promoters.
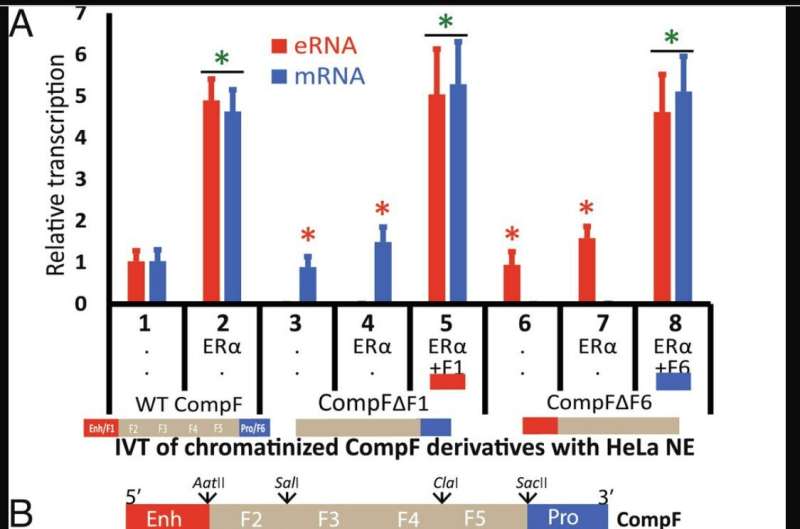
Enhancer-mediated regulation of gene expression has been mostly studied in intact living cells. "However, although much has been learned from these systems, it is difficult to control certain components in intact cells, limiting our mechanistic understanding of the process," Panigrahi said. "For this reason, we designed a cell-free assay that enables us to control the availability of different reaction components and to determine how this affects transcription."
"In the cell-free system, we saw that the enhancer and the promoter make close physical contact when the gene is transcribing, that is, making mRNA transcripts of the DNA sequence," Panigrahi said. "But we discovered that not only the gene but also the enhancer is being transcribed in the cell-free system, as happens in live cells."
Furthermore, they found that the transcription of the enhancer reflects the transcription of the promoter. "If we know the transcription status of the enhancer, we know the transcription status of the promoter and vice versa," Panigrahi said. "If we omit the promoter, then transcription of the enhancer is markedly reduced and vice versa. Enhancer and promoter transcription are tightly interconnected."
Previous studies using cell-based assays suggested that enhancer transcription somehow activated promoter transcription.
"What we are saying is that this goes both ways, not just one way," Panigrahi said. "Enhancer transcription activates promoter transcription and vice versa. Not only that, if the amount of transcription in the enhancer is reduced, then the promoter transcription is also reduced and vice versa. There is transcriptional interdependence between enhancers and promoters, which was not known before."
The researchers propose that such interdependence and regulatory specificity can be explained if the enhancer and the promoter are entangled within a transcriptional bubble that both provides shared resources for transcription and is regulated by the transcript levels generated.
"We are currently developing additional methodologies to conclusively test this transcriptional bubble model that enables entanglement of the participating enhancer-promoter pairs," said O'Malley, chancellor in the Department of Molecular and Cellular Biology and associate director of basic research at the Dan L Duncan Comprehensive Cancer Center at Baylor. "Our cell-free assay can be used to study promoter-enhancer interactions for any gene of interest, both in health and in disease."
More information: Anil K. Panigrahi et al, Enhancer–promoter entanglement explains their transcriptional interdependence, Proceedings of the National Academy of Sciences (2023).
Journal information: Proceedings of the National Academy of Sciences
Provided by Baylor College of Medicine

















