Surprising new mechanism of heat shock response identified in yeast cells

How cells respond in the face of stress is a critical component of maintaining homeostasis and preventing disease, but its mechanisms are poorly understood. New research out of the University of Chicago has fit together another piece of the puzzle by identifying a key protein, Sis1, involved in the rapid activation of the heat shock response, which allows cells to respond in the face of temperature changes that can lead to misfolded proteins. This has implications for conditions where changes in the stress response have been implicated in the pathology of the disease.The results were published on Jan. 4 in the Journal of Cell Biology.
In response to a sudden heat shock, cells activate a cascade of cellular activities to maintain protein integrity, necessary for maintaining cell function. This response is primarily mediated by the protein heat shock factor 1 (Hsf1). Activation of Hsf1 leads to the production of critical protein quality control factors that detect and address misfolded proteins, but it has been unclear how other chaperone proteins involved in the heat shock response affected the activation of Hsf1 and its subsequent effects. The team set out to investigate this question in yeast.
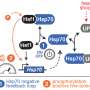
Initially, the investigators believed that the chaperone protein Hsp70 was likely the key to this process. Under ordinary circumstances, Hsp70 represses the activity of Hsf1; following heat shock, Hsp70 relocates to misfolded proteins and leaves Hsf1 free to activate protein quality control factors. In this way, Hsp70 might act like a "first responder," while Hsf1 gets to work sending out reinforcements to identify and clean up misfolded proteins. But the numbers didn't quite make sense.
"The thing that was confusing is that Hsf1 is a transcription factor, so it has to be located in the nucleus of the cell, but the previous model could not account for how rapidly the cell could then communicate between the cytoplasm—where the protein quality control factors could address misfolded proteins—and the nucleus," said co-first author Asif Ali, Ph.D., a postdoctoral scholar at UChicago.
The localization and concentration of Hsp70 didn't fit well with this model, so the team started to consider other candidates. This brought them to Sis1, a member of the Hsp40 protein family. Perhaps, they hypothesized, Sis1's relocation from the nucleus to the cytoplasm was the key to the puzzle. But even that wasn't so simple.
"When we did the experiment, I was initially very discouraged," said co-first author Zoe Feder, now a Ph.D. student at Harvard University. "We were expecting to see Sis1 completely leave the nucleus, but instead we saw it concentrated in these rings on the nucleolus. It looked weird and didn't make any sense, but it ended up being one of the most interesting aspects of the paper."
What they found was surprising: After a heat shock, rather than relocating to the cytoplasm (where it could start addressing misfolded proteins), Sis1 relocated instead to the edge of a cellular structure called the nucleolus, found inside the nucleus of the cell. This led to a sudden realization.
The nucleolus is the place where ribosomes are assembled in eukaryotic cells. Ribosomes are large and critically important, as they are responsible for the production of other proteins within the cell. Disruption of ribosomal integrity would have dramatic effects on protein production within the cell.
"We now think that the primary effect of heat shock is a local disruption of these ribosomal proteins," said senior author David Pincus, Ph.D., an Assistant Professor in the Department of Molecular Genetics and Cell Biology at UChicago. "What Sis1 and chaperone machinery might be doing is sequestering and degrading orphaned ribosomal proteins. By targeting the nucleolus, Sis1 doesn't have to leave the nucleus, but it does move away from Hsf1, allowing it to activate the heat shock response. This makes that response much faster than if Sis1 were leaving the nucleus entirely.
"This points toward a notion that hasn't been articulated in our field before," Pincus continued. "It's not just that the heat shock response targets unfolded proteins, but that it initially targets a very specific type of protein—these ribosomal proteins that are extremely physiologically important."
In essence, in response to heat shock, Sis1 quickly moves to the nucleolus, where it can interact with ribosomal proteins to begin repairing the damage. In the process, it breaks away from Hsf1, allowing Hsf1to activate and call for back up in the form of protein quality control factors.
These results could have implications for diseases like neurodegeneration and cancer, where changes in the stress response have been implicated in disease pathology.
"There's this idea that in cancer, cells have hijacked the heat shock response to sustain malignant growth," said Pincus. "On the other end, in Huntington's Disease, Hsf1 is degraded and the heat shock response is blocked, leading to protein aggregates. Sis1 may be a key regulatory node in this regulatory response—it may be a potent therapeutic handle if we can find a way to target it."
More information: Zoë A. Feder et al. Subcellular localization of the J-protein Sis1 regulates the heat shock response, Journal of Cell Biology (2020).
Journal information: Journal of Cell Biology
Provided by University of Chicago Medical Center